Ali Islam
RSNA Large Language Model Benchmark Dataset for Chest Radiographs of Cardiothoracic Disease: Radiologist Evaluation and Validation Enhanced by AI Labels (REVEAL-CXR)
Jan 21, 2026Abstract:Multimodal large language models have demonstrated comparable performance to that of radiology trainees on multiple-choice board-style exams. However, to develop clinically useful multimodal LLM tools, high-quality benchmarks curated by domain experts are essential. To curate released and holdout datasets of 100 chest radiographic studies each and propose an artificial intelligence (AI)-assisted expert labeling procedure to allow radiologists to label studies more efficiently. A total of 13,735 deidentified chest radiographs and their corresponding reports from the MIDRC were used. GPT-4o extracted abnormal findings from the reports, which were then mapped to 12 benchmark labels with a locally hosted LLM (Phi-4-Reasoning). From these studies, 1,000 were sampled on the basis of the AI-suggested benchmark labels for expert review; the sampling algorithm ensured that the selected studies were clinically relevant and captured a range of difficulty levels. Seventeen chest radiologists participated, and they marked "Agree all", "Agree mostly" or "Disagree" to indicate their assessment of the correctness of the LLM suggested labels. Each chest radiograph was evaluated by three experts. Of these, at least two radiologists selected "Agree All" for 381 radiographs. From this set, 200 were selected, prioritizing those with less common or multiple finding labels, and divided into 100 released radiographs and 100 reserved as the holdout dataset. The holdout dataset is used exclusively by RSNA to independently evaluate different models. A benchmark of 200 chest radiographic studies with 12 benchmark labels was created and made publicly available https://imaging.rsna.org, with each chest radiograph verified by three radiologists. In addition, an AI-assisted labeling procedure was developed to help radiologists label at scale, minimize unnecessary omissions, and support a semicollaborative environment.
Direct Multitype Cardiac Indices Estimation via Joint Representation and Regression Learning
May 25, 2017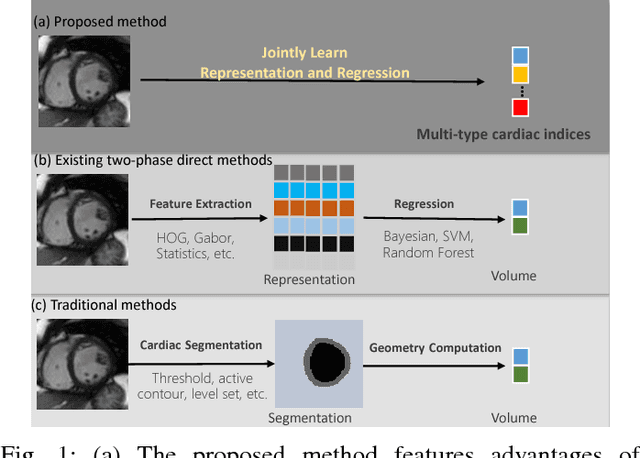
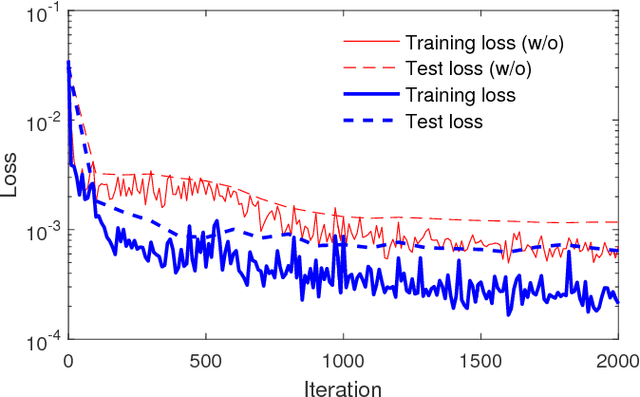
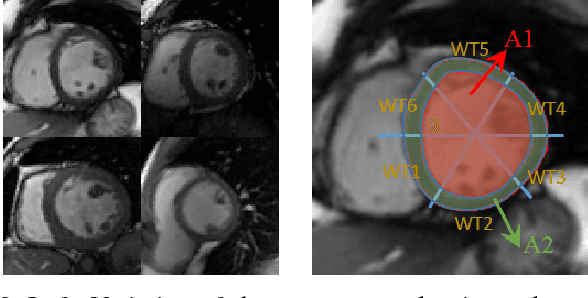
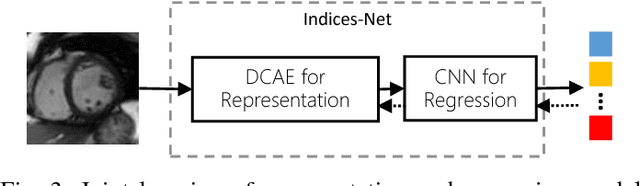
Abstract:Cardiac indices estimation is of great importance during identification and diagnosis of cardiac disease in clinical routine. However, estimation of multitype cardiac indices with consistently reliable and high accuracy is still a great challenge due to the high variability of cardiac structures and complexity of temporal dynamics in cardiac MR sequences. While efforts have been devoted into cardiac volumes estimation through feature engineering followed by a independent regression model, these methods suffer from the vulnerable feature representation and incompatible regression model. In this paper, we propose a semi-automated method for multitype cardiac indices estimation. After manual labelling of two landmarks for ROI cropping, an integrated deep neural network Indices-Net is designed to jointly learn the representation and regression models. It comprises two tightly-coupled networks: a deep convolution autoencoder (DCAE) for cardiac image representation, and a multiple output convolution neural network (CNN) for indices regression. Joint learning of the two networks effectively enhances the expressiveness of image representation with respect to cardiac indices, and the compatibility between image representation and indices regression, thus leading to accurate and reliable estimations for all the cardiac indices. When applied with five-fold cross validation on MR images of 145 subjects, Indices-Net achieves consistently low estimation error for LV wall thicknesses (1.44$\pm$0.71mm) and areas of cavity and myocardium (204$\pm$133mm$^2$). It outperforms, with significant error reductions, segmentation method (55.1% and 17.4%) and two-phase direct volume-only methods (12.7% and 14.6%) for wall thicknesses and areas, respectively. These advantages endow the proposed method a great potential in clinical cardiac function assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge